Study IDs why some TB bacteria prove deadly
Single mutation protects TB bacteria from antibiotics, immune assault
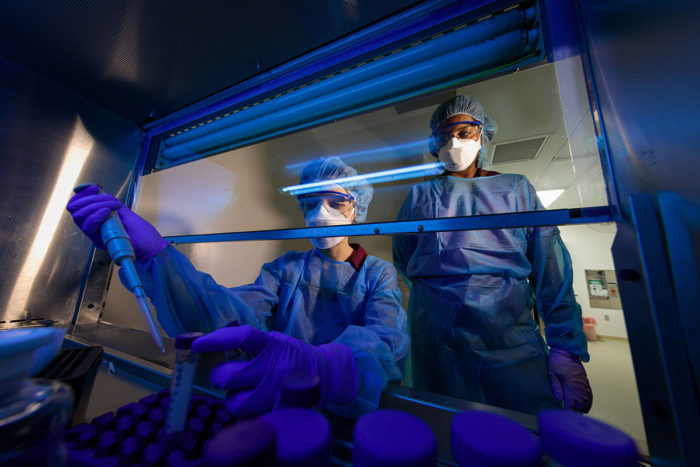 Matt Miller
Matt MillerGraduate student Nicole Howard works in a biosafety cabinet with the bacteria that cause tuberculosis (TB), while Shabaana Khader, PhD, looks on. Howard and Khader found that the same mutation that gives TB bacteria drug resistance also elicits a weaker immune response.
People who fall sick with drug-resistant tuberculosis (TB) face daunting odds. Only about two in three survive the illness, unlike people with drug-sensitive TB, of whom more than 90 percent survive.
Part of the reason drug-resistant TB is so lethal is because some antibiotics of first choice don’t kill the TB bacteria, forcing doctors to treat the infection with second-line medications that are often more toxic and less effective.
But the TB bacteria also may be undermining the body’s ability to defend itself. Researchers at Washington University School of Medicine in St. Louis have found that the same mutation that makes TB bacteria withstand a first-line drug also elicits a different – and probably weaker – immune response in mice.
“As the bacteria become drug resistant, they change physiologically and trick the immune system to behave differently,” said Shabaana A. Khader, PhD, an associate professor of molecular microbiology and the study’s senior author. “If we’re going to fight TB bacteria effectively, we’re going to have to understand what the drug-resistant bacteria are doing to the immune system to elicit protection.”
The findings are published Sept. 17 in Nature Microbiology.
There have been hints for years that drug-resistant bacteria might interact with the body differently than drug-sensitive strains. Anecdotal reports suggest that resistant TB bacteria cause more damage to the lungs and spread more widely through the body.
Khader, working with graduate student and first author Nicole Howard and colleagues, analyzed the immune response in mice infected with Mycobacterium tuberculosis, the bacteria that cause TB. They compared a strain that is resistant to all first- and many second-line antibiotics with a drug-sensitive strain of TB.
The mice’s immune systems battled the sensitive bacteria by releasing a powerful immune molecule known as IL-1 beta and ramping up the ability of their immune cells to burn sugar. Both actions are crucial for an effective immune assault on the TB bacteria.
But when infected with the drug-resistant strain, the mice failed to pull out the big guns. Their immune cells did not burn more sugar, and they failed to produce much IL-1 beta. Instead, they released a different immune molecule, IFN beta, which is associated with a feeble and sometimes detrimental immune response to TB.
“People always thought that development of drug resistance just meant that there’s a change in how bacteria respond to antibiotics,” Khader said. “But this shows that the whole immune environment is changing in ways that we haven’t been fully aware of.”
To find out why drug-resistant bacteria elicit a different immune response, the researchers scanned the whole genomes of several drug-resistant strains. They found that a change of one letter in the genetic code gave bacteria the ability to withstand rifampicin, an important first-line antibiotic, and also altered the immune response.
Treating TB is a long, drawn-out process. The bacteria grow slowly and are wrapped in a protective shell, so it takes a combination of four antibiotics given repeatedly over six months to eradicate them. Boosting the immune response would be like sending in fresh troops to a protracted battle – it could put an end to the fight, quickly and decisively. Clinical trials are already underway to find out whether immune-enhancing drugs combined with antibiotics improves outcomes for people with TB. But because of the inherent dangers of working with drug-resistant bacteria, nearly all research into strengthening the immune response has been conducted using drug-sensitive strains.
“If you’re going to do host-directed therapeutics, we need to know what immune pathways to target,” Khader said. “And these pathways may turn out to be different for drug-sensitive and drug-resistant TB.”
Howard and Khader’s findings suggest that the people most in need of better therapeutics – those infected with drug-resistant strains – may not benefit from immune enhancers that are designed based on drug-sensitive bacteria.
“We don’t know enough about the differences between resistant and sensitive TB to be confident that the therapeutics and vaccines we’re designing are going to work,” Khader said. “We’re going to have to do those studies.”